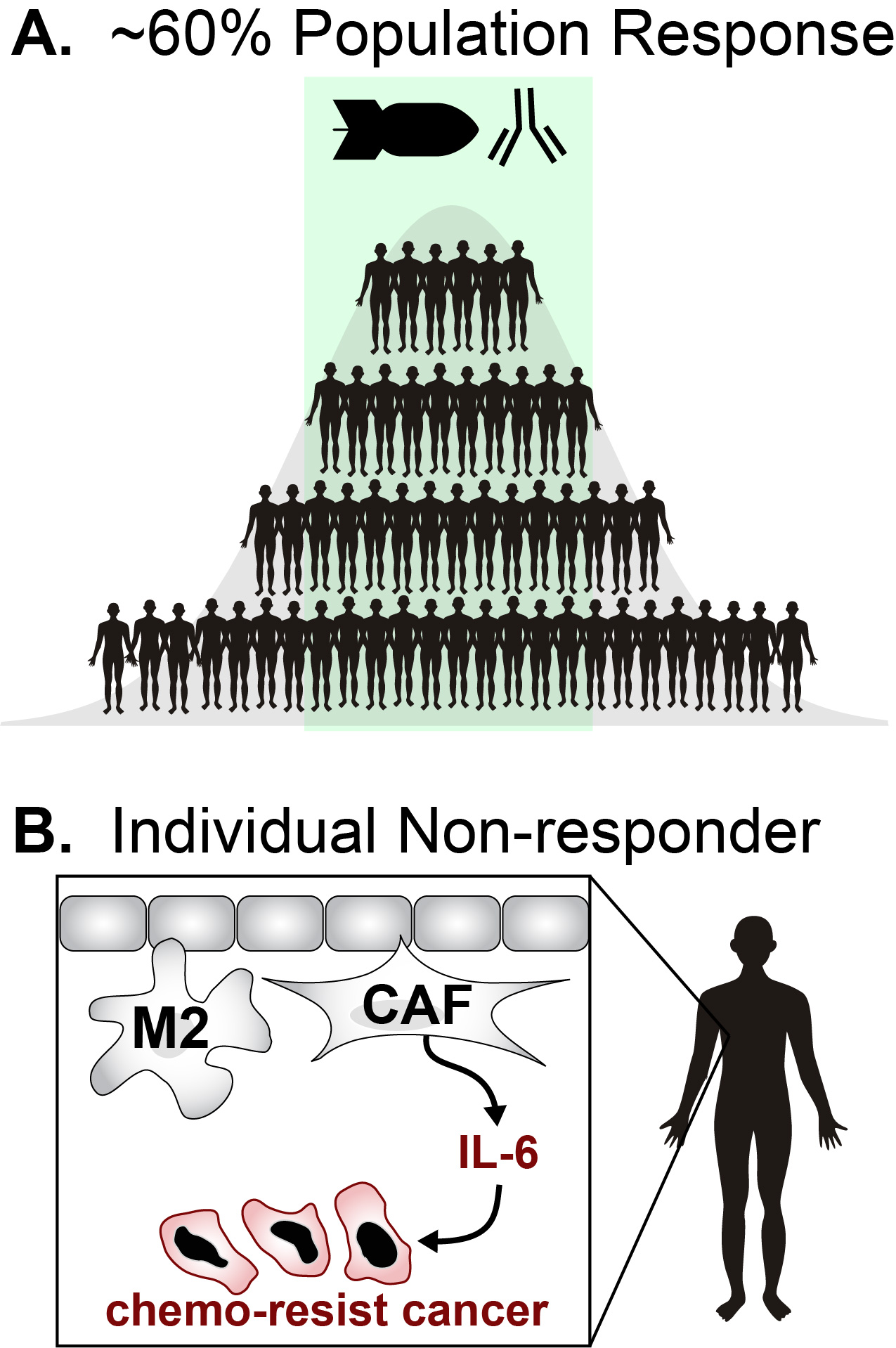
Recently, evidence of chemotherapy-Immunotherapy (chemo-IO) synergy was uncovered in a statistical analysis of 100 clinical trials (JAMA Netw Open., 2020, 3, e1920833). While exciting, this observation is largely empirical as we do not understand the mechanisms behind improved response rates. More generally, it is difficult to define the patient populations where this synergy will manifest itself. For example, for triple-negative breast cancer (TNBC), new chemo-IO combinations help most (~60%) but nevertheless to fail many (Figure 1A). In fact, recent guidelines have stated that a “one size fits all” approach is likely insufficient:
- “Given the heterogeneity of breast cancer, even when restricted to HER2-negative disease, it is also possible that “one size will never fit all” and there is no best treatment for most patients.” (J Clin. Oncol, 2014, 32, 3307)
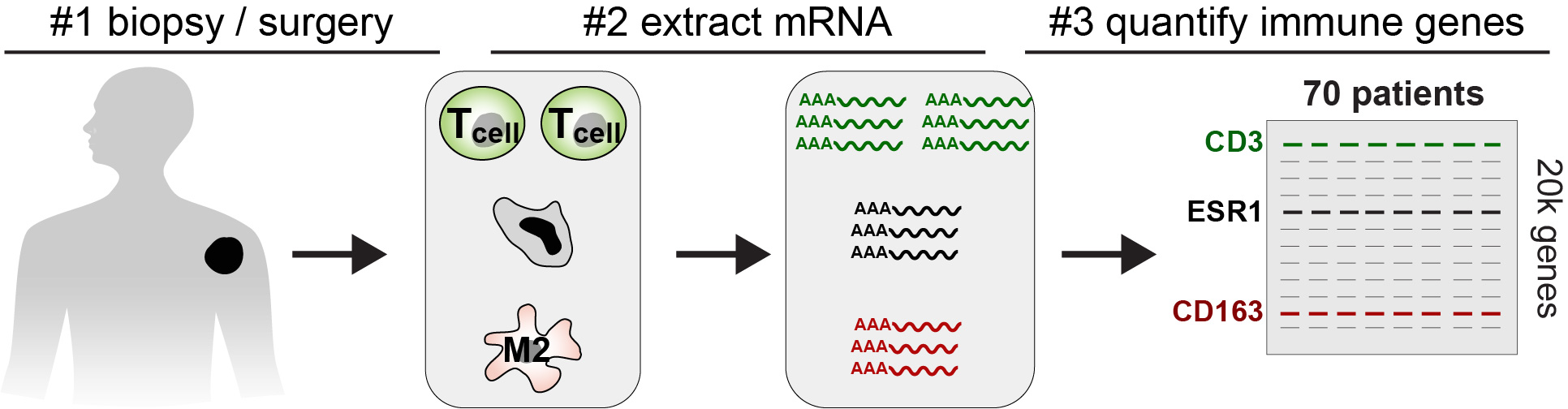
One major source of heterogeneity is infiltrating immune and stromal cells that can directly effect resistance (Figure 1B). For example, it has been shown that macrophage and fibroblast-derived interleukin-6 (IL-6) can drive chemo-resistance to anthracyclines and platinum agents (Figure 1B). In addition, anthracyclines can selectively deplete myeloid derived suppressor cells and anti-inflammatory T-cells. Critically, tumor-RNAseq can be used infer the types of immune cells within a tumor microenvironment based on the differential expression of immune-cell genes (Figure 2). CIBERSORT has become the standard method for estimating frequencies of immune cells from bulk tumor-RNAseq across a wide range of cancers. In addition, its inferences have been shown to correlate with histological subtypes and have shown predictive power for both drug-response and patient-survival.
In collaboration with oncologists at Emory, we develop related algorithms to better understand the immune-cancer battlefield within primary TNBC samples.